If The Dna Template Had The Base Sequence Agct, What Would Be The Mrna Base Sequence?
Genetic code
Our genes are encrypted books conveying the secrets of life. In order to sympathize these secret messages, we would demand to know the code and utilize the aforementioned set of rules, in contrary, to decode it. In this commodity, we'll take a closer await at the genetic code, which allows DNA and RNA sequences to be "decoded" into the amino acids of proteins.

How do our cells brand proteins – Transcription and Translation
Our genes are written every bit the nucleotide base of operations pairs (A, T, G, C) in the Dna. For a gene to exert its function, the genetic information must read out to build a protein. This process is chosen gene expression.
There are 2 steps for making proteins from genes:
Beginning, inside the nucleus, a process that makes copies of a certain factor in the form of massager RNAs (mRNAs), called transcription.
Second, these mRNAs are exported exterior of the nucleus to the cytoplasm for ribosomes to brand polypeptides/ proteins. This step is called translation.
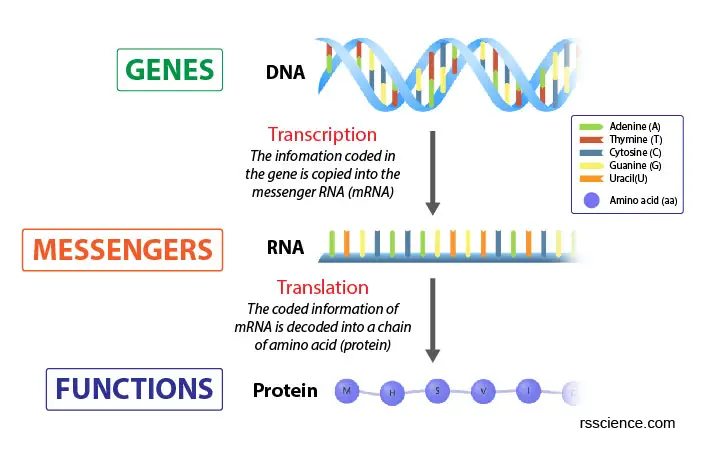
[In this paradigm] The Fundamental Dogma of Biological science.
Genes contain the information to build proteins that maintain cell viability. This building process is done in ii steps: Transcription and Translation.
The re-create from DNA to RNA is simple: post-obit the complementary base pairing dominion. In Dna, there are 4 nitrogenous base options: Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). Each base can but bond with one other, A with T and C with G. This is called the DNA complementary base pairing rule.
To transcript DNA into mRNA, the dominion is the same. The but difference is that Uracil (U) replaces Thymine (T). So, Chiliad ↔ C, A → U, and T → A. In our cell, the transcription is done past an enzyme called RNA polymerase in the nucleus, which can synthesize mRNA from a Dna template.
Dna to mRNA: Using complementary base pairing rules
Knowing this dominion, you lot tin figure out the complementary strand to a single DNA strand based only on the base pair sequence. For instance, let's say you know the sequence of one Dna strand that is every bit follows:
Dna (coding strand): v'-TTG ACG ACA AGC TGT TTC-iii'
Using the complementary base pairing rules, you tin conclude that the complementary strand is:
DNA (template strand): 3'-AAC TGC TGT TCG ACA AAG-v'
RNA strands are also gratuitous with the exception that RNA uses U instead of T. Therefore, you lot tin likewise infer the mRNA strand that would be produced from the get-go DNA strand. Information technology would be:
mRNA: 5'-UUG ACG ACA AGC UGU UUC-3'
RNA to Protein: Using genetic codons
While DNA (genes) and RNA (messengers) use similar codes fabricated of 4 units, proteins are built very differently. Proteins are built using 20 units called amino acids. Translating mRNA to protein becomes much more complicated. To guide this translation, cells follow the genetic code. Co-ordinate to the genetic code, the genetic information is organized in triplets of nucleotides, and each triplet is translated into ane amino acrid.
For instance, the mRNA above will translate into
Protein: Leu – Thr – Thr – Ser – Cys – Phe
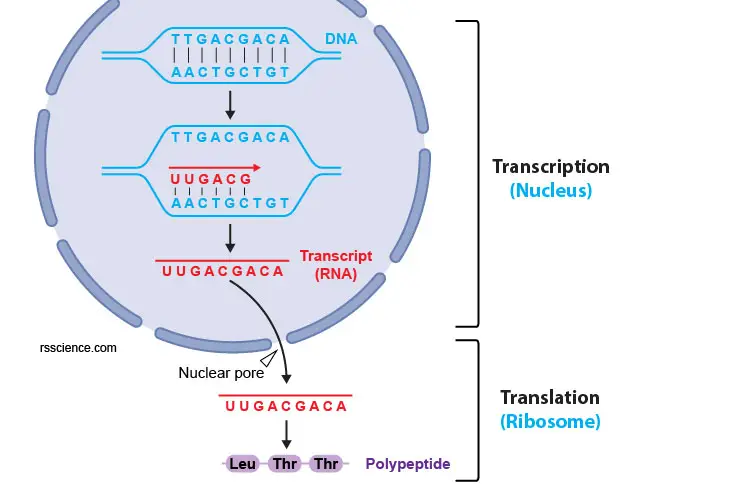
[In this figure] The process of gene expression. From a gene to a protein, there are two steps, transcription and translation. The Dna needs to exist transcripted to mRNA using complementary base pairing (i.e., A pairs with U; T pairs with A; C pairs with Thou; G pairs with C). Next, the mRNAs are exported to the cytoplasm through nuclear pores and are translated to proteins by ribosomes.
Note: A brusque concatenation of amino acids is often referred to as a "polypeptide". When the number of amino acids adds upward (ordinarily > xxx units) and the polypeptide chain folds into a 3D structure, we call it a "protein".
At that place are three features of codons:
- Each codon specifies an amino acid. The full set of relationships betwixt codons and amino acids is summarized equally a Condon Chart or Table.
- One "Start" codon (AUG) marks the outset of a poly peptide. AUG encodes the amino acid, called Methionine.
- Three "Cease" codons marking the cease of a protein and finish the translation.
Who tin read these codes? Ribosome as a decoding machine
Codons in an mRNA are read by a ribosome during translation. A ribosome is a particle-like prison cell organelle made of ribosomal RNA (rRNA) and ribosomal proteins. A ribosome consists of two major components: the small and large ribosomal subunits. Three binding sites for tRNA (A, P, and E sites) between the two subunits. Read more about ribosomes.
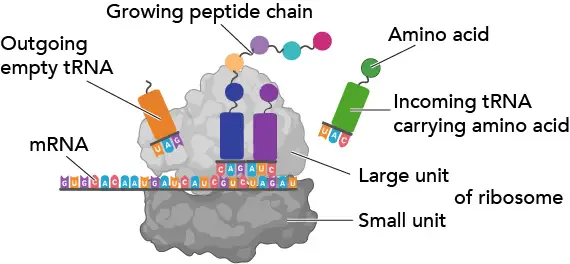
[In this figure] Ribosome.
Ribosomes work like decoding machines to translate the code sequence of mRNA into a poly peptide. Scientists similar to phone call ribosomes, the molecular micro-machines, to admire how exquisite the ribosomes' design is!
Transfer RNA (tRNA)
The transfer RNA (tRNA) is one type of RNA molecule. Its job is to carry the amino acid that matches the mRNA codon to the ribosome.
The tRNA contains a 3-letter code on ane side and carries a specific amino acid on the other side. The lawmaking on tRNA (called an anticodon) must match the three-letter code (the codon) on the mRNA already in the ribosome. The item amino acid that tRNA carries is determined by a three-letter of the alphabet anticodon it bears. For example, if the three-alphabetic character code is AUG on mRNA, the tRNA that carried Methionine (Met) will be selected and recruited to the ribosome. This is an essential part of the translation process, and information technology is surprising how few "errors of translation" occur.
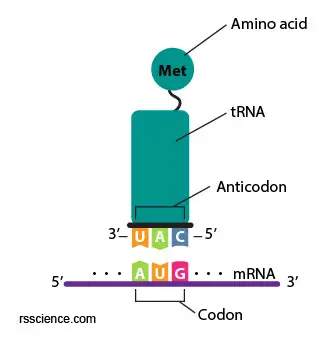
[In this figure]A anticodon UAG on the tRNA matches to the AUG on the mRNA (complimentary) and bring the right amino acrid (Methionine) to the ribosomes.
Protein translation begins with a start codon (always AUG → Methionine) and continues until a stop codon (any 1 of the three: UAA, UAG, or UGA) is reached. mRNA codons are read from 5′ end to 3′ stop, and its order specifies the order of amino acids in a protein from N-terminus to C-terminus.
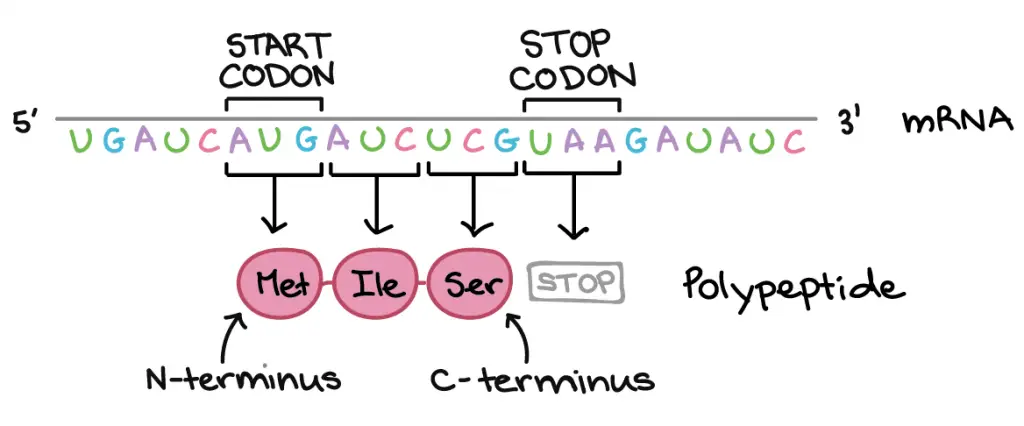
[In this figure]Directionality: Deoxyribonucleic acid and RNA read from 5' end to iii' end. Instead, proteins or polypeptides read from Northward-terminus (amino group) to C-terminus (carboxyl group). The beginning and the cease of a translation is marked by the Start and Stop codons, respectively.
Photograph credit: khanacademy
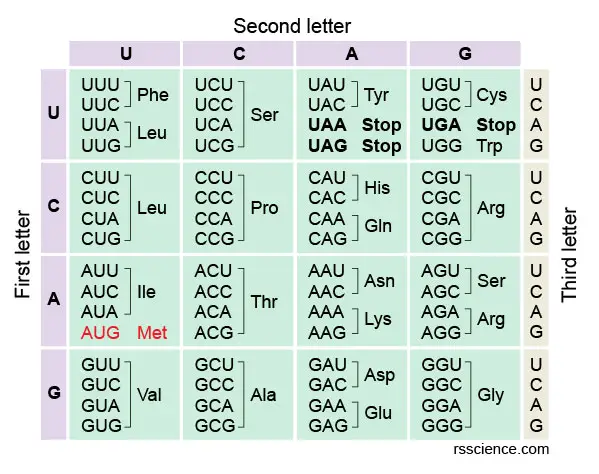
The amino acids codon chart
The full set of relationships between codons and amino acids (or stop signals) is called the genetic code. The genetic code is often summarized in a codon chart (or codon table), where codons are translated to amino acids.
[In this image] Condon set tin also be presented as a codon wheel.
Photo credit: wiki
How practice you read the codon nautical chart?
The codon chart may look intimidating at commencement. In fact, information technology is not difficult at all once you empathise its dominion.
Let'south take codon ACU every bit an instance. If you want to know which amino acrid ACU encodes, you get-go wait at the left side of the table. Find the "A" on the centrality of the left side, which refers to the first letter of the codon triplet. All these codons starting with "A" are in this row.
Side by side, we look at the pinnacle of the table. This upper axis indicates the 2d letter of the codon triplet. Once we find "C along the upper axis, it tells us about the column in which our codon volition be plant. Find the intersecting box of "A" row and "C" column in the tabular array. You will see this box containing four codons and easily find the 1 you're looking for.
In our example, ACU encodes Thr (or Threonine). You may also discover that all ACU, ACC, ACA, and ACG encode the same amino acid. Notice that many amino acids are represented in the table by more i codon. For example, in that location are half dozen unlike ways to "write" Leucine in the language of mRNA (see if you lot can find all half-dozen).
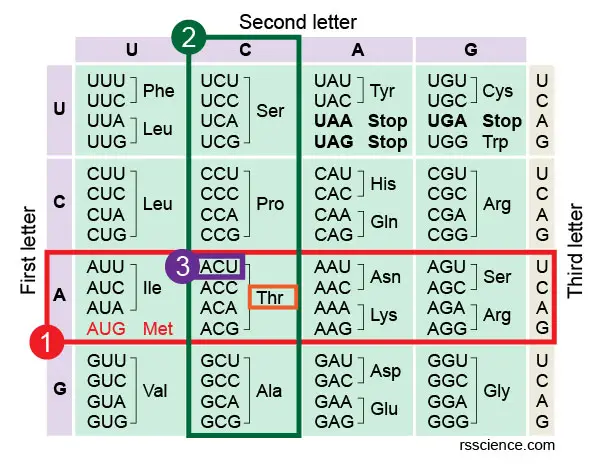
[In this prototype] How to read the amino acids codon nautical chart?
Following Footstep 1-iii to notice the codon triplet in the table.
In this tabular array, you can also run across that UAA, UAG, and UGA do not encode any amino acrid, meaning they are stop codons.
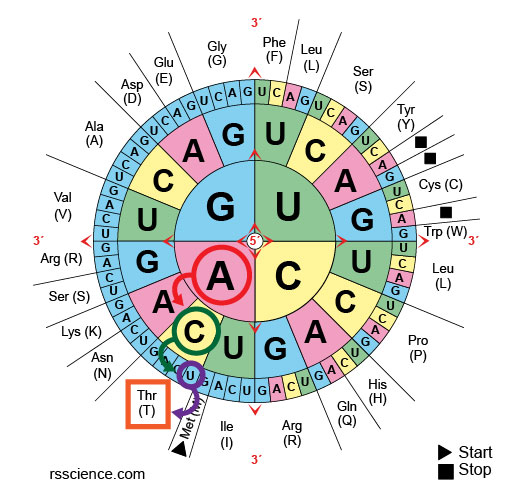
[In this prototype] For a codon wheel, the dominion is the aforementioned: start from the center to find the kickoff letter of the alphabet of triplet, then move toward the periphery for 2nd and 3rd letters.
You and your family or classroom can play the "Codon Bingo" to get familiar with the genetic lawmaking. Here is a downloadable version.
Reference Tabular array: a summary of all amino acids codons
| Amino Acid | Codon |
| Phenylalanine (Phe) | UUU, UUC |
| Leucine (Leu) | UUA, UUG, CUU, CUC, CUA, CUG |
| Methionine (Met) / Outset Codon | AUG |
| Valine (Val) | GUU, GUC, GUA, GUG |
| Serine (Ser) | UCU, UCC, UCA, UCG, AGU, AGC |
| Proline (Pro) | CCU, CCC, CCA, CCG |
| Threonine (Thr) | ACU, ACC, ACA, ACG |
| Alanine (Ala) | GCU, GCC, GCA, GCG |
| Tyrosine (Tyr) | UAU, UAC |
| Histidine (His) | CAU, CAC |
| Glutamine (Gln) | CAA, CAG |
| Asparagine (Asn) | AAU, AAC |
| Lysine (Lys) | AAA, AAG |
| Aspartic Acrid (Asp) | GAU, GAC |
| Glutamic Acid (Glu) | GAA, GAG |
| Cysteine (Cys) | UGU, UGC |
| Tryptophan (Trp) | UGG |
| Arginine (Arg) | CGU, CGC, CGA, CGG, AGA, AGG |
| Glycine (Gly) | GGU, GGC, GGA, GGG |
| Isoleucine (Ile) | AUU, AUC, AUA |
| Terminate Codon | UAA, UAG, UGA |
Molecular structures of Amino acids
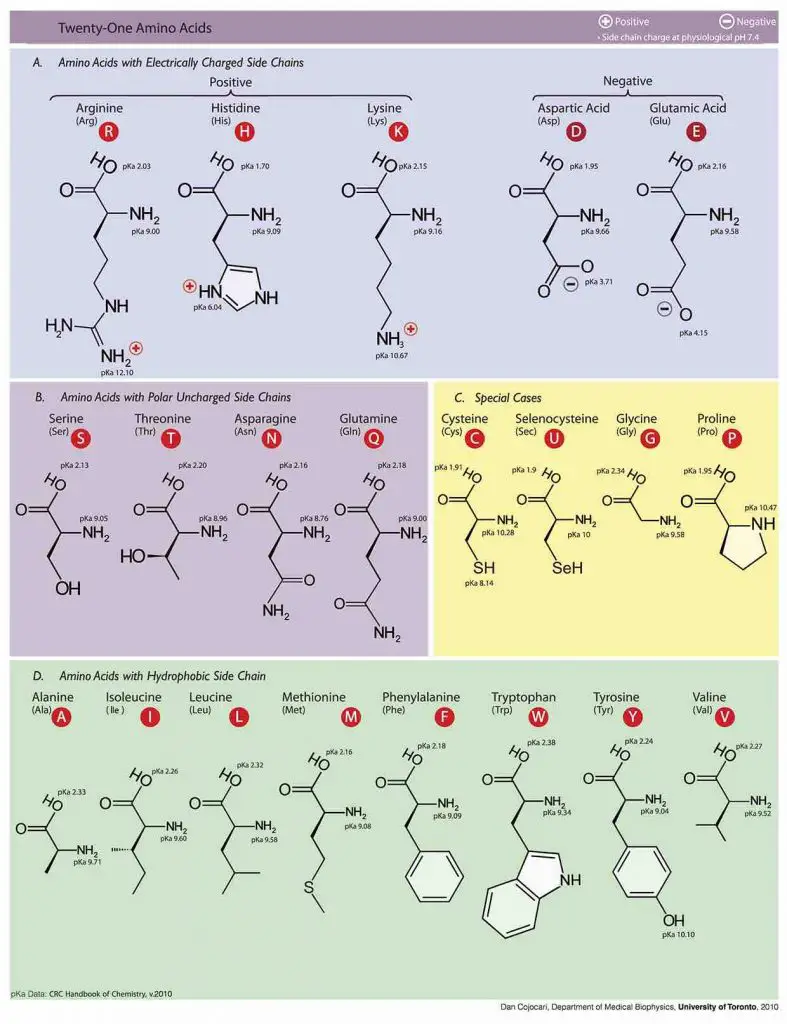
Photo source: wiki
Standard Genetic Code
The genetic code we mentioned hither is universal; with merely a few exceptions, virtually all species (from bacteria to humans) apply the aforementioned set up of standard code. Some ciliates, such every bit Paramecium bursaria, use unusual genetic code.
Some other exception is mitochondrial Dna. Mitochondria have their ain copies of DNA likewise as an contained system of ribosomes and tRNAs. If you are not familiar with mitochondria, click here to learn more than about mitochondria.
The mitochondrial lawmaking is slightly unlike from the standard genetic lawmaking. Moreover, dissimilar species accept their own versions of mitochondrial codes. For example, our (vertebrate) mitochondrial code is dissimilar from the ane yeast uses. AGA and AGG encode Arginine (Arg) in the standard genetic code. Nevertheless, AGA and AGG act as finish codons in the vertebrate mitochondrial code. In addition, UGA and AUA change from terminate codon and Isoleucine (Ile) to Methionine (Met) and Tryptophan (Trp), respectively, in mitochondria.
The aforementioned situation also happens in the plant's chloroplast and plastid codes.
Codon usage biases
Although near organisms use the standard code, however, they may take their own biases in terms of choosing which codons to use. For instance, baking yeasts prefer using UGU for Cysteine. In contrast, in human cells, we adopt UGC.
Codon usage biases could be the event of natural selection (tRNA affluence). For laboratories to produce certain proteins in a big quantity, researchers may perform "codon optimization" to resynthesize genes in such a fashion that their codons are more than advisable for the desired expression host (i.e., making human proteins in E coli. bacteria).
What is reading frame?
Since the Dna sequence is read by triplets, starting from which letter (or reading frame) becomes a disquisitional problem.
Permit'due south await at an example. The mRNA below can be translated into three totally different orders of amino acids, depending on the frame in which information technology'south read. How practise our cells know which of these proteins to make?
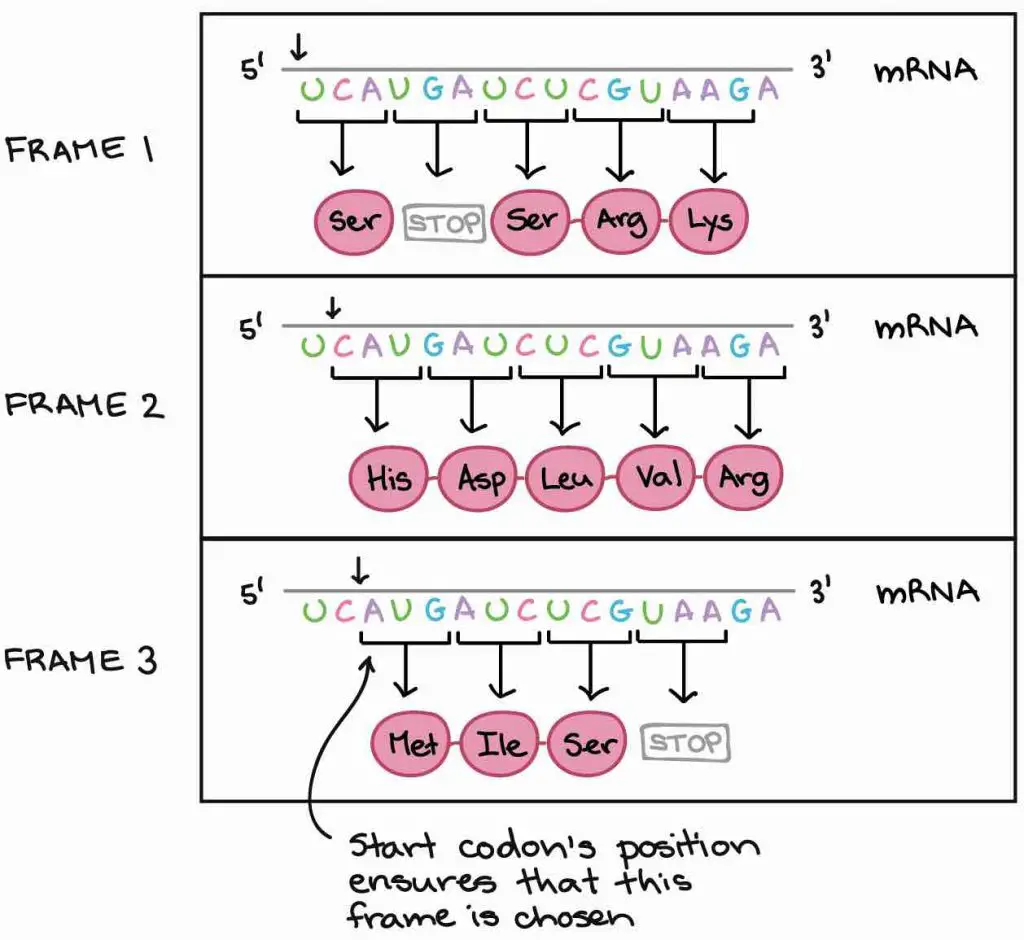
[In this image] Iii possible reading frames could lead to totally dissimilar results.
Photo credit: khanacademy
Our cells utilise a very smart strategy to solve this trouble – the "showtime codon". Because the translation only begins at the start codon (AUG) and continues in successive groups of three, the position of the start codon ensures that the mRNA is read in the right frame (in the example in a higher place, in Frame 3).
What happens if the Deoxyribonucleic acid sequences are messed upward – Mutation
Mutations (changes in Deoxyribonucleic acid sequences) may derail the genetic information and crusade cells to make the incorrect proteins. Mutations are the major cause of cancers and many genetic disorders.
Even a single base pair contradistinct (called point mutation) can cause a significant consequence. Betoken mutations can have 1 of 3 effects.
Silent mutation
Offset, the base commutation can be a silent mutation where the contradistinct codon corresponds to the same amino acid. For example, changing from UCU to UCC has no effect since both codons equally encode Serine (Ser).
Missense mutation
2nd, the base substitution can be a missense mutation where the contradistinct codon corresponds to a dissimilar amino acid. For example, changing from UCU to UGU volition turn Serine (Ser) to Cysteine (Cys). If this mutation happens in the critical region (i.e., enzymatic site) of the protein, a point mutation can mess upwardly the whole poly peptide part.
Nonsense mutation
3rd, the base substitution can exist a nonsense mutation where the altered codon becomes a stop signal. This is the worst cause considering the translation volition end besides early, resulting in a truncated protein.
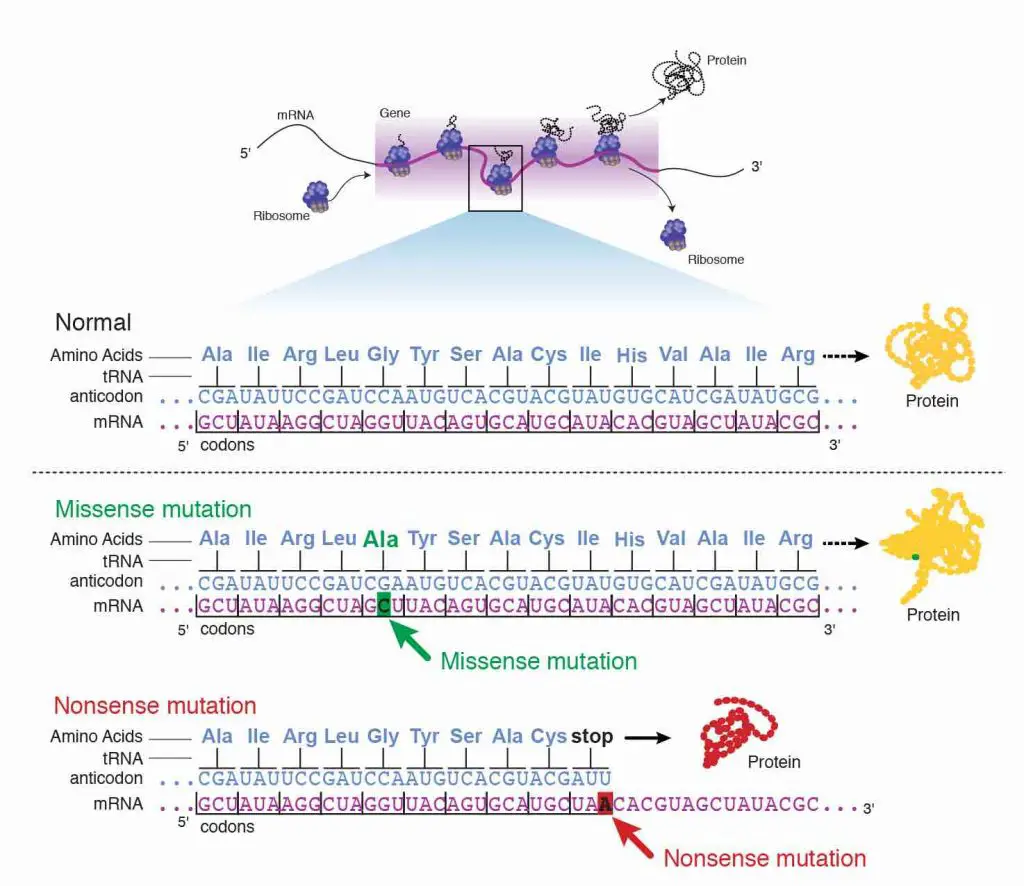
[In this image] The examples of showing the result of missense mutation and nonsense mutation.
Photo credit: NIH
Mutations could too happen when nucleotides are inserted or deleted from the original Dna sequence. The insertion or deletion of "one or two" nucleotides can alter the reading frame (frameshift mutation). A frameshift can totally mess upwards the amino acid orders "downstream" the mutation site.
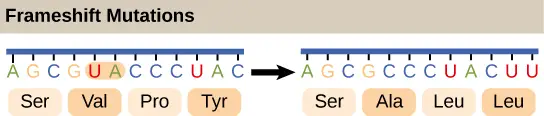
[In this image] The example of showing the consequence of frameshift mutation.
Photo credit: openstax
How was the genetic code discovered?
The understanding of genetic code is the foundation of modem biotechnology. Without the power to read the DNA data, many heady techniques and therapies, including personalized medicine, gene therapy, CRISPR cistron editing, and recombinant protein drugs, won't be.
To crack the genetic code, researchers needed to effigy out how nucleotides sequences in a DNA or RNA molecule could encode the sequence of amino acids. In the mid-1950s, physicist George Gamow predicted that the genetic code is probable composed of triplets of nucleotides – considering the possible combination of duplet is not enough (four×4 = 16), and that of quadruplet is as well many (4x4x4x4 = 256), to cover 20 kinds of amino acids.
The actual experiments to pinpoint the genetic code began in 1961 past American biochemist Marshall Nirenberg. Nirenberg was able to link the relationships between nucleotide triplets to particular amino acids by 2 experimental innovations:
- He can synthesize artificial mRNA molecules with specific, known sequences.
- He had a organisation to translate mRNAs into polypeptides outside of a jail cell (a "cell-complimentary" system). Nirenberg did so in a test tube of cytoplasm from burst E. coli leaner, which contains all the ingredients needed for translation.
Nirenberg started with an mRNA molecule consisting merely of the nucleotide uracil (called poly-U). When he added poly-U mRNA to the prison cell-costless organization, he found that the polypeptides made consisted exclusively of the amino acid – Phenylalanine (Phe). Nirenberg concluded that UUU might code for phenylalanine. Using the same approach, he discovered triplet CCC codes for Proline (Pro).

Photo credit: khanacademy
Following this concept, the biochemist Har Gobind Khorana extended Nirenberg's experiment by synthesizing artificial mRNAs with more complex sequences. By 1965, Nirenberg, Khorana, and their colleagues had deciphered the unabridged genetic code.
For their contributions, Nirenberg and Khorana (forth with another genetic code researcher, Robert Holley) received the Nobel Prize in Physiology or Medicine in 1968.

[In this paradigm] The Nobel Prize in Physiology or Medicine 1968 was awarded jointly to Robert W. Holley, Har Gobind Khorana, and Marshall Due west. Nirenberg "for their interpretation of the genetic code and its function in protein synthesis."
Photo credit: The Nobel Prize
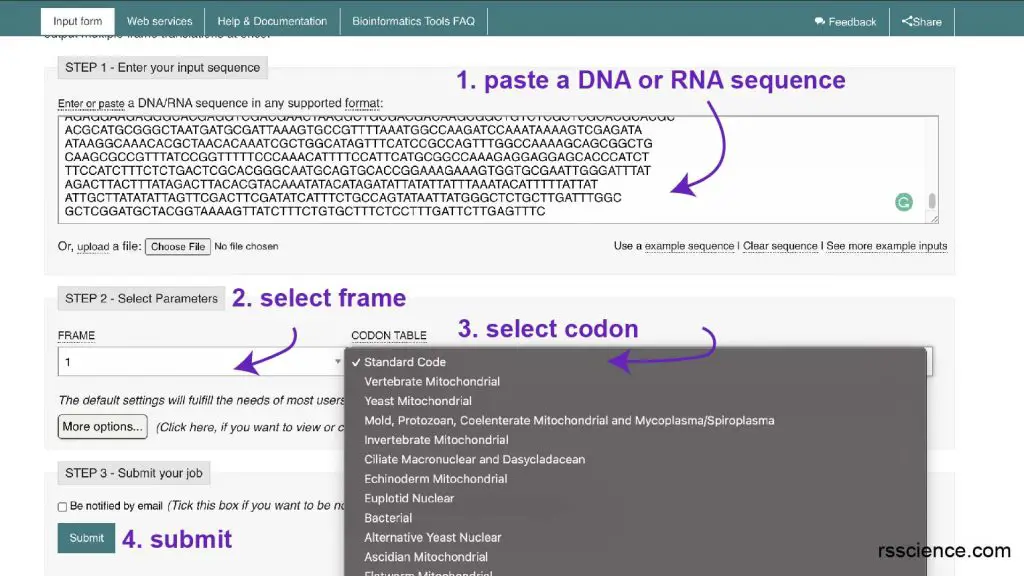
In my research that I need to clone a particular DNA for protein expression, I typically utilize EMBOSS Transeq from EMBL-EBI.
Step 1: Paste a piece of Dna sequence, you can use their example sequence.
Step 2: Select the reading frame yous want to use
Step 3: Select codon. I typically apply standard code.
Step 4: Hit submit
Step five: Wait for the protein sequence result!
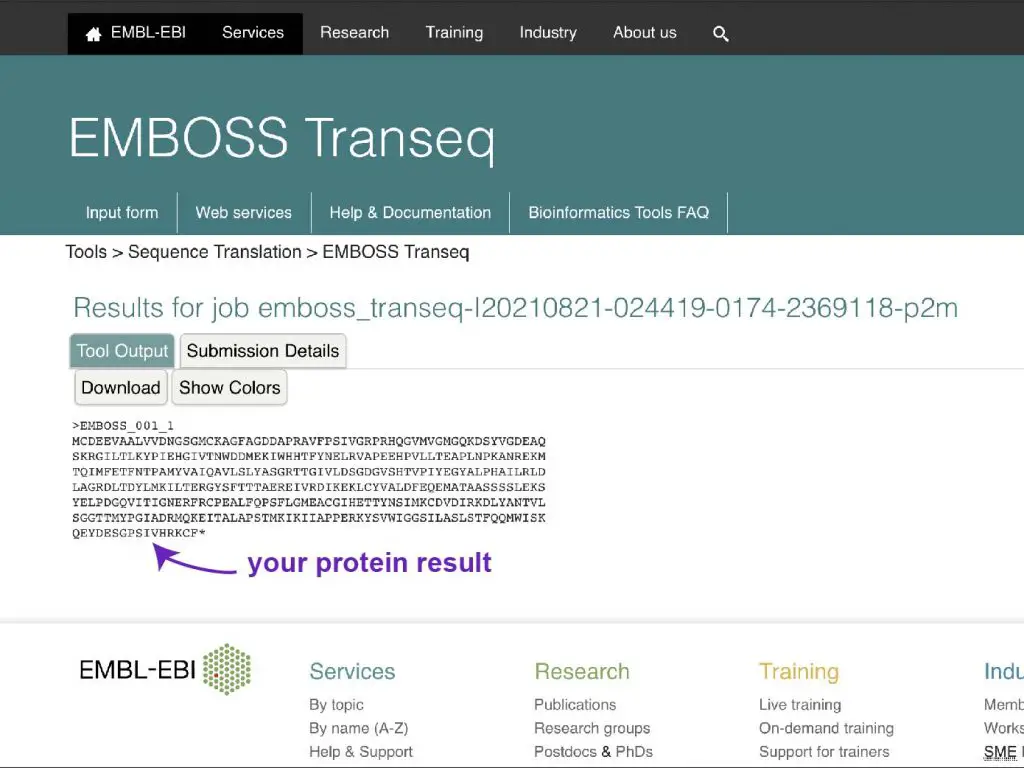
References
If The Dna Template Had The Base Sequence Agct, What Would Be The Mrna Base Sequence?,
Source: https://rsscience.com/codon-chart/
Posted by: rivasforengs.blogspot.com


0 Response to "If The Dna Template Had The Base Sequence Agct, What Would Be The Mrna Base Sequence?"
Post a Comment